DNA Methylation: What’s the Difference Between 5mC and 5hmC?
DNA methylation is perhaps the most studied epigenetic mechanism, known for its role in gene regulation and development. Related DNA modifications, once considered mere intermediates in the demethylation process, are now recognized for their stability and biological significance. 5-hydroxymethylcytosine, the first product of demethylation, plays an important role in the mammalian brain and development. Here, we discuss these two distinct DNA modifications and the methods used to identify and quantify them across the genome.
DNA Methylation
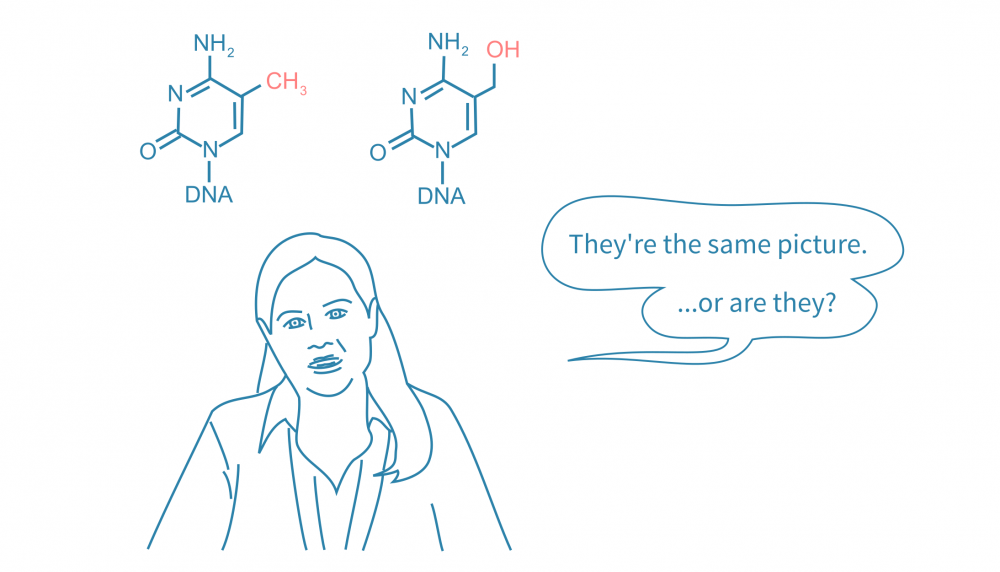
DNA methylation is an epigenetic mechanism, where a methyl group is added to the cytosine to form 5-methylcytosine (5mC). It typically occurs at CpG sites and plays key roles in regulating gene expression, chromosomal stability, and genomic imprinting. During development, patterns in DNA methylation across the genome change as cells differentiate, resulting in distinct methylomes in different cell types by the end of development.
Cytosine Demethylation
A methylated cytosine can be demethylated in response to various factors such as heat stress, nutrition, or infection. DNA demethylation can play a role in differential regulation of gene expression and occurs through either active or passive mechanisms. Passive demethylation happens when DNA methylation is not maintained during cell division and DNA replication. Active demethylation is mediated by ten-eleven translocation enzymes (TETs), which oxidize 5mC stepwise to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC).
5-Hydroxymethylcytosine
While 5mC has been extensively studied and implicated in disease development and pathways, 5hmC was once thought to be just an intermediate in active demethylation. However, several studies from the past fifteen years have shown that 5hmC has an important role during mammalian development and in the nervous system. Aberrant 5hmC has been associated with various neurodevelopmental disorders and cancer. Characterising and understanding the role of 5hmC has become important to understand neurodevelopment and disease. Importantly, 5hmC has distinct effects compared to 5mC.
Profiling 5mC and 5hmC
To profile DNA methylation, it's necessary to distinguish between unmethylated and methylated cytosines using sequencing or microarray methods. The most common technique is sodium bisulfite treatment, which deaminates unmethylated cytosines, leaving methylated ones untouched. The unmethylated cytosines are then converted into thymines through PCR. However, this method does not differentiate between 5mC and 5hmC, as both are resistant to deamination. As a result, studies using sodium bisulfite identify both 5mC and 5hmC as methylated sites with no distinction between the two.
Telling the two apart: Oxidative Bisulfite Treatment
The first method developed to identify 5hmC was oxidative bisulfite sequencing (OxBS-seq). This technique adds an oxidative step to convert 5hmC to 5fC. Since 5fC is sensitive to bisulfite treatment, these cytosines are deaminated, allowing researchers to distinguish them from 5mC. To identify CpGs with 5hmC positions, the standard bisulfite treatment must be run in parallel with the oxidative protocol, and the sequencing or microarray data is compared site by site. While this method allows the identification of both 5mC and 5hmC, it has the drawback of requiring parallel sequencing or microarray runs, which increases costs.
Newer Methods
Recent advances have led to the development of more efficient methods to read 5mC and 5hmC:
- CMD1-Deaminase Sequencing (CD-seq) and CMD1-TET Bisulfite Sequencing (CT-seq): 5mC modification enzyme 1 (CMD1) converts 5mC to 5-glyceryl-methylcytosine (5gmC) without affecting 5hmC. CT-seq can also convert 5hmC into thymine, leaving 5mC unaffected.
- Chemical-assisted Pyridine Borane Sequencing Plus (CAPS+): This method selectively oxidizes 5hmC to 5fC, and further oxidation converts 5fC to 5caC, allowing highly sensitive, quantitative sequencing of 5hmC.
- NTET-assisted Pyridine Borane Sequencing (eNAPS): In this technique, 5mC is oxidized by eNTET to 5fC and 5caC, enabling precise mapping of 5mC.
- Oxford Nanopore Technologies Sequencing: Nanopore sequencing has significantly advanced the direct detection of 5mC and 5hmC.
Read more
Contact us
Leave your email address here with a brief description of your needs, and we will contact you to get things moving forward!

